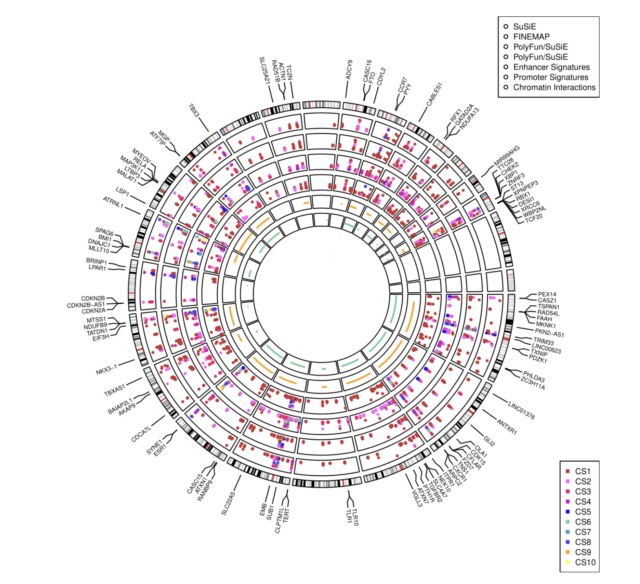
Fine-scale mapping of breast cancer susceptibility loc
Breast cancer remains a major health challenge worldwide, with its genetic underpinnings still not fully understood. To bridge the gap between genetic susceptibility and disease mechanisms, advanced computational approaches are necessary. Dr. Kyriaki Michailidou and her team at the Cyprus Institute of Neurology and Genetics aim to leverage the MeluXina supercomputer to implement fine-scale mapping of breast cancer susceptibility loci, advancing our understanding of the genetic architecture of the disease.
The Challenge of this Research
The primary challenge in breast cancer genomics lies in identifying true causal variants from the thousands of associations discovered through genome-wide association studies (GWAS). Current challenges include:
1. Large-scale datasets (consisting of millions of variants): Requiring large computational resources and capabilities
2. Noisy Data: The genetic data is vast and includes many variants that may not be causally related to the disease but rather indirect associations to the trait of interest.
3. Limited Causal Insights: GWAS identify regional associations rather than variants with direct effects on the phenotype. Therefore, GWAS do not provide clear causal relationships between variants and disease.
4. Complex Genetic Architecture: Breast cancer risk is influenced by numerous low-penetrance variants, making it difficult to pinpoint specific causal variants.
The MeluXina Solution
To address these challenges, the project employs the high-performance computing capabilities of MeluXina. The approach includes:
1. Bayesian Fine-Mapping: Using PolyFun, SuSiE, and FINEMAP, the project prioritizes variants based on functional annotations and computes posterior probabilities of causality.
2. Independent Signal Identification: GCTA-COJO will be used to identify independent genetic signals within loci.
3. Computational Resources: Utilizing 10,000 node hours on MeluXina, with an average of 512 threads and 1024 GB of total job memory.
Impact
The impact of this research is multifaceted:
1. Enhanced Identification of Causal Variants: By fine-mapping over 200 breast cancer-associated regions, the project aims to significantly improve the identification of causal variants.
2. Improved Risk Estimation: Incorporating identified causal variants into polygenic risk scores will enhance breast cancer risk estimation and patient management.
3. Broader Application: The methodologies developed can be applied to other common cancers and complex traits, broadening the impact of the research beyond breast cancer.
Conclusion
This project represents a significant step forward in breast cancer genomics. By utilizing the computational power of MeluXina, Dr. Michailidou’s team aims to refine fine-mapping techniques, providing deeper insights into the genetic factors contributing to breast cancer. The anticipated outcomes will not only advance scientific knowledge but also improve clinical risk assessments and patient care strategies, setting a new benchmark for future genetic research in cancer and other complex diseases.